Introduction
Materials and Methods
Plant materials
Electrolyte leakage assay
Gene expression study
In vitro ubiquitination assay
Subcellular localization
Results
Phenotypic evaluation and electrolyte leakage analysis of both genotypes under chilling treatment
Expression analysis of OsCRF1 in Donganbyeo and Satbyeolbyeo
OsCRF1 RING domain has E3 ubiquitin ligase activity
Subcellular localization of the OsCRF1 protein
Discussion
Introduction
Cold stress, which includes chilling (0-15°C) and/or freezing (< 0°C), limits the spatial distribution and seasonal growth pattern of crops, and adversely affects plant growth and development (Zhu et al., 2007; Adam and Murthy, 2013). Rice (Oryza sativa L.) is a major staple crop and feeds over half the world’s population (Zhang et al., 2011); however, this crop is highly vulnerable to cold stress, especially during the reproductive stage (Broad et al., 1980). When rice plants are exposed to chilling stress, seedlings exhibit slow growth, withering, yellowing, and reduced tillering in vegetative stage (Ye et al., 2009). In addition, chilling stress causes delayed heading, inhibition in panicle opening, poor pollen growth, pollen sterility, and reduces the number of spikelet as well as the panicle area during the reproductive stage (Jacob and Pearson 1999; Andaya and Mackill 2003). Gene regulation under chilling stress is well-understood in typical temperate crops such as barley, wheat, rye, among others (Chinnuisamy et al., 2007). However, some subtropical cereal crops such as rice, maize, and sorghum are considered as being highly chilling-sensitive compared to temperate crops, suggesting that such plants exploit cold stress regulation mechanisms.
Generally, plants perceive cold signals through their membranes and various transduction elements, stimulating the transcription of several genes accordingly. In Arabidopsis, many transcription factors, which facilitate cold or freezing stress signaling and control the expressions of cold regulators, have been identified (Chinnusamy et al., 2010), and their homologs have been reported in other plants (Lissarre et al., 2010). It has been revealed that genes encoding the transcription factors ICE1, HOS1, ESK1, and CBF2 are involved in cold stress response (Chinnusamy et al., 2003; Dong et al., 2006; Xin et al., 2007; Novillo et al., 2008). To date, one of the most studied cold pathways for cold stress acclimatization by plants is the ICE-CBF-COR signaling pathway (Wang et al., 2017). In addition, several cold related genes have been isolated and characterized in Arabidopsis and other cold tolerant plants (Wang et al., 2017).
In higher plants, ubiquitin molecules bind to target proteins and regulate a variety of post-translational modifications. Three types of enzyme, ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (E2), and ubiquitin ligase (E3) are required to enable the cellular reactions mediating ubiquitination (Vierstra, 2009). RING E3 ligases usually interact with target protein(s), which are gradually attached to target(s) by ubiquitin molecules. They can be tagged with one or multiple ubiquitin molecules, each present in mono-multiple or polyubiquitination forms. Polyubiquitinated substrate proteins are usually modulated by the 26S proteasome for protein degradation (Smalle and Vierstra, 2004). The number and diversity of RING E3 ligases confer specificity to the ubiquitination pathway. According to subunit composition and mode of action, four E3 types have been reported in plants: RING/U box, homology to E6APC terminus, F-box protein (SCF), and a complex of Skp1. In Arabidopsis, more than 1300 genes are predicted to encode E3 ligases and among these, at least 469 genes are expected to encode RING domain-containing proteins (Stone et al., 2005). However, the role of RING finger proteins in modulating the response to cold stress via the ubiquitin/proteasome pathway has not yet been studied in depth.
RING E3 ligases play an important role in the adaptation to different environmental stresses through a variety of mechanisms such as signal transduction, hormone biosynthesis, or transcriptional factors (Lyzenga and stone, 2011). For example, the overexpression of rice E3 ligase encoding gene, OsCTR1, was linked to an increased tolerance to drought stress in Arabidopsis (Lim et al., 2014). Overexpression of Oryza sativa heat- and cold-induced 1 gene (OsHCI1) conferred tolerance to heat stress conditions in Arabidopsis (Lim et al., 2013). In addition, the heterogeneous overexpression of a gene encoding salt-induced rice RING finger protein (OsSIRP1) reduced the tolerance toward salinity stress (Hwang et al., 2016). Moreover, the overexpression of Oryza sativa heat tolerance at seedling stage gene (OsHTAS), encoding RING E3 ligase, enhances heat tolerance (Liu et al., 2016).
Previously, Lim et al. (2013) evaluated the expression patterns of 47 O. sativa RING finger protein genes (OsRFPs) in response to different abiotic stresses including salt, drought, heat, and cold stress. In this study, we selected one chilling-induced RING finger protein gene, named O. sativa chilling induced RING finger protein 1 (OsCRP1). In brief, we evaluated the phenotypic differences between two rice cultivars, Donganbyeo and Satbyeolbyeo, and investigated OsCRP1 expression patterns in both plants under chilling stress. Furthermore, we attempted to determine the subcellular localization of this gene in rice protoplasts and N. benthamiana, and examined the E3 ligase activity via In vitro ubiquitination assay.
Materials and Methods
Plant materials
Rice seedlings (O. sativa L. Donganbyeo and O. sativa L. Satbyelbyeo) were grown in mesh supported plastic containers filled with a half-strength Murashige and Skoog (MS) solution or in pots filled with a commercial soil (Baroker, Seoul Agricultural Materials Co., Korea), in a growth chamber (16/8 h light/dark photoperiod at 25°C with 70% relative humidity), for gene expression study under chilling treatment and electrolyte leakage assay, respectively. Two weeks-old plants grown in the plastic containers were treated with chilling stress (9°C) for 12 days. Stress-treated and non-treated leaves were harvested at 0, 1, 5, and 12 days.
Electrolyte leakage assay
To evaluate the cold tolerance capacity of Donganhyeo and Satbveolbyeo, we carried out electrolyte leakage assays on rice plants grown at 25 and 9°C for 12 days. Samples were collected at various time intervals: 0, 1, 3, 5, 7, 9, and 10 days. Disks were punched from leaves with fully developed rosettes, transferred into tubes containing 8 mL of water, and incubated for 24 h. The tubes filled with leaf disks and water were autoclaved, and each ion leakage percentage value was calculated before and after this process. To evaluate reproducibility three biological replicates were performed for each treatment.
Gene expression study
To study gene expression levels, total RNA was isolated with TRIzol® Reagent according to the manufacturer's protocol (Life Technologies, Carlsbad, CA). For quantitative RT-PCR (qRT-PCR) assay, the primer pair for OsCRF1 (Os04g43300) was designed using the Beacon Designer 8 software (Beacon Designer 8, Premier Bio soft). qRT-PCR assay was conducted as follows; all reactions contained 10 µL of SYBR Green TOPrealTM qPCR 2× PreMIX (Enzynomics, Daejeon, Korea), 1 µL of each gene-specific primer (10 pmol/µL), and 1 µL of cDNA in a final volume of 20 µL. The fluorescent signals were detected using the CFX Connect™ RT-PCR Detection System (Bio-Rad, Hercules, CA, USA). OsActinI (Os03g50885) was used as the internal control (Park et al., 2014). Three biological replicates were tested for each sample, and the qRT-PCR was run using following program: 95°C for 10 min, followed by 50 cycles of 95°C for 10 s, 58–60°C for 30 s, and 72°C for 30 s, with a final extension step at 72°C for 10 s. After completion of the qRT-PCR assay, relative expression levels were calculated using the 2–DDCt algorithm Schmittgen 2001 (Livak and ). The primers used in this study are listed in Table 1.
In vitro ubiquitination assay
To perform the In vitro ubiquitination assay, the RING-domain coding sequences of OsCRF1 (OsCRF1-RD) were amplified with Pfu Turbo DNA polymerase (Stratagene, La Jolla, CA), using the RING domain-specific primers. The PCR products were then cloned into a pMAL-c5X vector (New England BioLabs, Ipswich, MA, USA), and transformed into an E. coli strain (DH5α). Then, the recombinant MBP-OsCRF1-RD and MBP-empty plasmids were expressed in E. coli strain BL21 (DES) pLys (Promega, Madison, WI, USA) and purified using affinity chromatography in an amylose resin (New England Biolabs). The full-length coding sequence of AtUBC10 (E2) was cloned into the pET-28a (+) vector (Novagen, Gibbstown, NJ, USA) with 6X His tag. The clone was also expressed in E. coli strain BL21 (Promega) and purified by Ni-NTA Purification System (Invitrogen Life Technologies, Carlsbad, CA). In vitro ubiquitination assay was carried out according to Hwang et al. (2017) with some modifications. Each purified MBP-OsCRP1-RD (300 ng) or non-recombinant MBP (negative control) was mixed with 50 ng yeast E1 (Boston Biochemicals, Cambridge, MA, USA), 250 ng of purified AtUBC10 (E2), 10 mg bovine Ub (Sigma-Aldrich, St. Louis, MO, USA), along with 20X reaction buffer (1 M Tris, pH 7.5; 40 mM ATP; 100 mM MgCl2; and 40 mM DTT). The reaction mixtures were incubated in a 37°C shaker for 3 h. To stop the reaction, 5X SDS sample buffer was added, followed by boiling at 100°C for 5 min. The boiled samples were run on a 10% SDS-PAGE gel and transferred onto a nitrocellulose membrane (DoGen, Seoul, Korea). Immunoblot analysis was carried out using anti-ubiquitin (Sigma-Aldrich, St. Louis, MO) as the primary antibody and goat anti-rabbit IgG peroxidase as secondary antibody (Sigma-Aldrich). To detect the poly-ubiquitinated product, the WESTSAVE STARTM substrate (AB Frontier, Seoul, Korea) was used according to the commercial manual. Images were captured using the ChemiDocTM XRS+ imaging system (Bio-Rad, Hercules, CA).
Subcellular localization
To isolate rice protoplasts, seedlings were grown in a plastic container with 1/2 Murashige and Skoog (MS) medium for 14 days. Leaves of 14 days-old seedlings were chopped and immersed in an enzyme solution [1% cellulase R-10 (Yakult Honsa Co. Ltd., Tokyo, Japan), 0.25% macerozymeR-10 (Yakult Honsa), 0.1% BSA, 10 mM MES, 500 mM mannitol, 1 mM CaCl2] for 6 h with gentle shaking (Zhang et al., 2011). Protoplast pellets were collected by centrifugation for 4 min at 300 g after filtration through a 40- µm. nylon mesh. The protoplasts were suspended in W5 solution (154 mM NaCl, 125 mM CaCl2, 5 mM KCl, 5 mM glucose, 1.5 mM MES) and incubated by gentle shaking for 10 min, followed by incubation in ice for 5 h. Subsequently, the solution was resuspended in MaMg solution (0.4 M mannitol, 15 mM MgCl2, 4.7 mM MES). To determine the subcellular localization of OsCRF1, the full-length gene was amplified by pfu turbo DNA polymerase (Stratagene, La Jolla, CA) and then, the PCR product was inserted into the multiple cloning sites (MCS) of the enhanced yellow fluorescent protein (EYFP) vector. The constructed vector was transfected into the isolated protoplasts using a polyethylene glycol (PEG) solution (40% PEG, 400 mM mannitol, 100 mM Ca(NO3)2) for 30 min at room temperature 24-25°C. Finally, the PEG solution was diluted in W5 solution, followed by resuspension of the protoplasts and incubation overnight at room temperature. Fluorescence images were obtained using a multiphoton confocal laser scanning microscope (model LSM 510 META; Carl Zeiss, Jena, Germany) at the Korea Basic Science Institute Chuncheon Center.
To confirm the subcellular localization, a transient expression assay was also conducted in tobacco leaves. The 35S: OsCRF1-EYFP or 35S: EYFP genes were transformed in A. tumefaciens, followed by incubation in 25 mL of YEP solution supplemented with 50 mg/L kanamycin at 28°C overnight. Then, the samples were continuously washed 3–4 times with an infiltration buffer (10 mM MgCl2, 10 mM MES, pH 5.6), before adding 200 mM acetosyringone to each sample. p19 protein was added to the prepared samples and injected into the tobacco leaves. The signals were also observed using the multiphoton confocal laser scanning microscope
Results
Phenotypic evaluation and electrolyte leakage analysis of both genotypes under chilling treatment
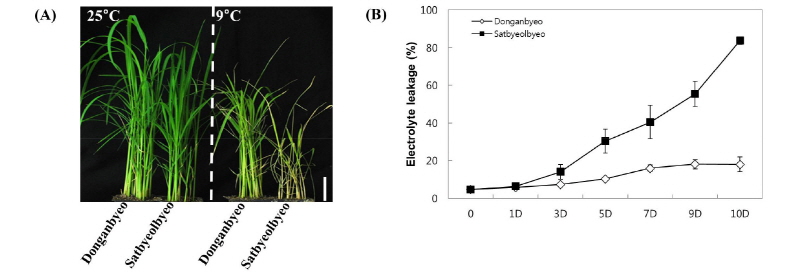
We observed the phenotypic differences between both cultivars (Donganbyeo and Satbeolbyeo) under normal conditions (25°C) and chilling stress (9°C) for 12 days. While similar phenotypes were observed among both cultivars under normal conditions, the Donganbyeo cultivar showed higher tolerance than Satbyeolbyeo (Fig. 1A) under chilling treatment. To examine leave damage under chilling treatment, the percentages of electrolyte leakage were measured for both cultivars through a time-course experiment carried out after the treatment. We found that the electrolyte leakage percentage significantly increased in Satbyeolbyeo until day 10; however, Donganbyeo showed less than 20% leakage 12 days after treatment (Fig. 1B), suggesting that Donganbyeo is more tolerant than Satbyeolbyeo under chilling stress.
Expression analysis of OsCRF1 in Donganbyeo and Satbyeolbyeo
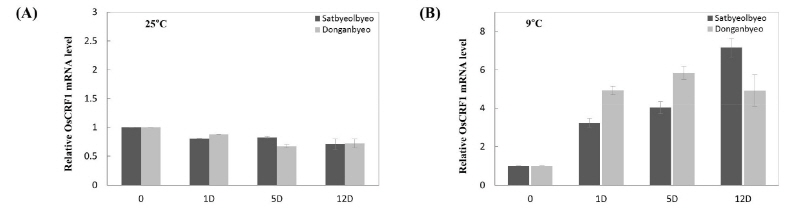
Previously, Lim et al. (2013) studied the expression patterns of 47 Oryza sativa RING finger protein genes in response to various abiotic stresses. Among them, we selected one gene (Os04g43300), which was determined as being highly induced under chilling stress in Donganbyeo (data not shown) using semi-quantitative RT-PCR (named OsCRF1). To examine the relative mRNA expression levels of OsCTR1 in both plants, qRT-PCR analysis was performed on both normal and chilling stressed leaf samples at different time points 0, 1, 5, and 12 days (Fig. 2). No significant difference was observed between both cultivars under normal conditions; however, the transcript level was significantly higher in Donganbyeo after 5 days under chilling stress. In contrast, the transcript level of Satbyeolbyeo became higher than that of Donganbyeo at day 12 (Fig. 3B).
OsCRF1 RING domain has E3 ubiquitin ligase activity
It is believed that RING domains confer E3 ligase activity and are involved in various cellular processes (Lim et al., 2013). We found that the OsCRF1 protein contains a HC-type RING domain, a PHD_SF domain, and two BRCT domains via NCBI Conserved Domain Search (www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) (Fig. 3A). OsCRF1 is composed of 629 amino acids and is predicted to have a molecular mass of 68.29 kDa. To investigate whether OsCRF1 possesses E3 ligase activity, we performed an In vitro ubiquitin assay on MBP-OsCRF1. However, the full-length protein was not well-expressed in E. coli (data not shown). Therefore, only the RING domain was cloned into the MBP vector. The purified MBP-OsCRF1-RD was mixed with ubiquitin reaction components, E1 (yeast), E2 (AtUBC10), Ub, and ATP, and subsequently incubated for 3 h at 30°C. Immunoblot analysis detected high molecular mass (polyubiquitin chain) of MBP-OsCRF1-RD with anti-Ub antibodies after incubation with all the reaction components (Fig 3B lane 4). However, no polyubiquitin chains were detected when MBP-OsCRF1-RD was incubated in absence of E1 or E2 (Fig. 3B, lanes 2 and 3). This result confirms that OsCRF1 harboring RING domain possesses E3 ubiquitin ligase activity.
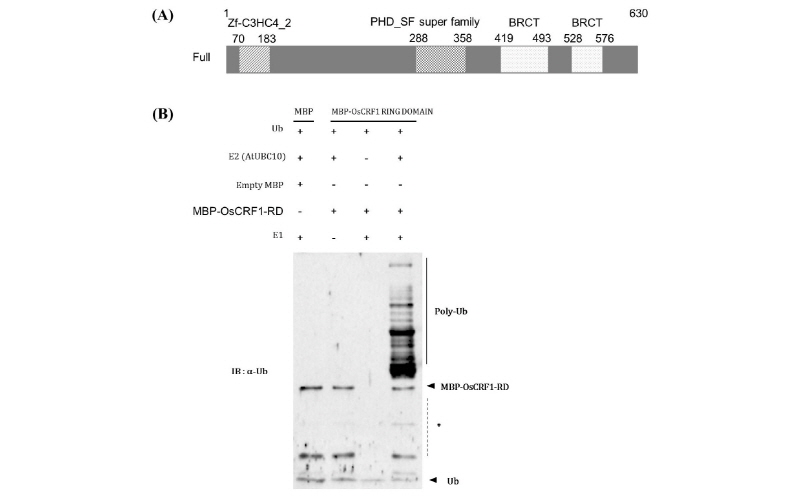
Fig. 3. Schematic diagram of OsCRF1 and ubiquitin assay. (A) The schematic diagram of OsCRF1 including HC-type RING domain (70-183 aa), PHD_SF super family (288-358 aa), and two BRCT domains (419-494 and 528-576 aa) (B) E3 ligase activity of OsCRF1. MBP-tagged OsCRF1 fusion protein was analyzed for E3 ligase activity in the presence (+) and/or absence () of E1 (yeast), E2 (AtUBC10), or E3.
Subcellular localization of the OsCRF1 protein
To determine the subcellular localization of OsCRF1, full-length OsCRF1 gene was fused with the enhanced yellow fluorescent protein (EYFP) vector in the C-terminal region, amplified into the CaMV 35S promoter, and transfected into rice protoplasts. The fluorescence signal of cto OsCFR1 was clearly observed in the cell nucleus, whereas the 35S: EYFP signal used as the control was found in both the nucleus and cytosol. To further confirm the OsCRF1 localization, the 35S: EYFP and 35S: OsCRF1-EYFP fusion proteins were transiently expressed in tobacco leaves. The 35S: OsCRF1 protein was distinctly localized in the nucleus (Fig. 4), supporting our former results.
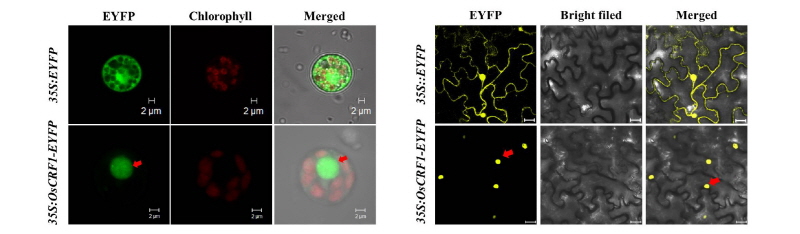
Fig. 4. Subcellular localization of the OsCRF1-EYFP fusion protein. (A) 35S: EYFP and 35S: OsCRF1-EYFP fusion proteins were transiently expressed in rice protoplasts. Green represents the fluorescence color of EYFP and red represents the fluorescent signal of chlorophyll. The arrow indicates the fluorescence of OsCRF1-EYFP. (B) 35S: EYFP and 35S: OsCRF1-EYFP fusion proteins were transiently expressed in tobacco. Yellow represents the fluorescence color of EYFP. The arrow indicates the fluorescence of OsCRF1-EYFP.
Discussion
Plants are constantly exposed to various environmental stresses, and low temperature (freezing or chilling) constitutes a major factor influencing plant physiological and morphological characteristics. Plants have developed a variety of mechanisms to enhance tolerance to low temperature stress, also called cold acclimation. However, the molecular machinery behind cold acclimation and freezing tolerance phenomena has mainly been studied in Arabidopsis and cold-season cereals. Cold response mechanism is a very complex trait, which involves many different metabolic pathways, gene regulations, and cell compartments (Hanan et al., 2005). Previously, several studies suggested that cold-regulated gene expression plays an important role in chilling stress tolerance and cold acclimation in plants (Zhu et al., 2007; Miura et al., 2013). Therefore, understanding cold stress related regulatory mechanisms seem paramount toward improving plant tolerance against low temperature stresses through genetic engineering.
In this study, we provide insights on the molecular functions of a chilling-induced RING finger protein isolated from rice. Previously, Lim et al. (2013) observed that OsCRF1 (originally named OsRFPHC12) was down-regulated upon cold stress (4°C). Here, we found that this gene was highly expressed in both phenotypes under chilling stress, suggesting that different regulation mechanisms of the gene might exist for either cold or chilling stresses; however, further investigations are needed to confirm this hypothesis. We assessed the phenotypic effect on two varieties and examined expression levels of OsCRF1 under chilling stress. The degree of cell membrane injury caused by chilling is reflected by the intracellular electrolyte leakage rate; therefore, the evaluation of electrolyte leakage was employed to determine the metabolic activity of Donganbyeo and Satbyeolbyeo during chilling treatment. Higher electrolyte leakage was observed in Satbyelbyeo than Donanbyeo, suggesting that the metabolic activity of Donanbyeo is higher than that of Satbyeolbyeo under chilling stress. Similarly, the overexpression of OsNAC5 in Arabidopsis enhanced its tolerance to cold, and OsNAC5 overexpressed-plants exhibited lower electrolyte leakage than wild-type ones (Song et al., 2011). The expression pattern of OsCRF1 was highly induced in both varieties during chilling stress, but lower expression was observed in normal conditions (Fig. 2B), suggesting the role of OsCRF1 in response to chilling stress. Interestingly, we found that the transcript level of OsCRF1 was higher in Donganbyeo compared with Satbyeolbyeo until 5 days; however, expression level of OsCRF1 was decreased at 12 days in Donganbyeo. It might be due to that regulation of protein accumulation and for maintaining cell balance; therefore, Donganbyeo plants might show tolerance phenotype. In addition, Satbyeolbyeo showed relatively low expression level of OsCRF1 at early stage and this initial damage might affect the subsequent growth, therefore Satbyeolbyeo plants might show sensitive phenotype (Fig. 1A). These results suggest that early high-expression of OsCRF1 might play a crucial role in conferring chilling tolerance in Donganbyeo. However, further work is required to confirm this hypothesis.
26S proteasome-mediated ubiquitin-dependent protein degradation is known to be an effective response to environmental stress (Smalle et al., 2003; Kurepa et al., 2008). Previously, we demonstrated the roles of several RING E3 ligases in perception and transduction under various abiotic stress conditions through ubiquitin-mediated protein degradation (Lim et al., 2014, 2015; Park et al., 2015; Hwang et al., 2017). In this study, we confirmed that OsCRF1, composed of 629 amino acids with HC-type RING domain, possesses E3 ligase activity via In vitro ubiquitination assay.
Previously, it was reported that the overexpression of RING finger E3 ligase high expression of osmotically responsive gene 1 (HOS1) confers increased sensitivity during low temperature stress, and that the degradation of HOS1-mediated substrate protein led to attenuated freezing stress responses in Arabidopsis through the protein ubiquitination degradation pathway (Dong et al., 2006). Similarly, a couple of studies demonstrated that the nucleus-localized genes act as cold-induced transcription regulators of genes in response to cold stress, such as ICE1 (inducer of CBF expression 1), an upstream transcription factor which regulates the transcription of CBF genes in response to low temperature stress (Chinnusamy et al., 2003). We found that OsCRF1 was localized in the nucleus (Fig. 4), suggesting that the E3 ligase might act as a regulator of nuclear protein expression under chilling stress. However, further studies would be necessary to shed a light on the molecular functions of OsCRF1 E3 ligase, including the development of OsCRF1-overexpressed transgenic plants, as well as mining and degradation assays on substrate proteins.
In conclusion, we observed a high expression of OsCRF1 under chilling stress, E3 ligase activity, and nucleus targeting, providing some important clues toward understanding the molecular mechanisms governing the response to chilling stress mediated by OsCRF1 pathways.
Moreover, based on the results of expression pattern of OsCRF1 in Donanbyeo and Satbyeolbyeo under chilling stress suggests that OsCRF1 exhibits its function in the weaning stage. However, further work is necessary to clearly understand the mechanism by which OsCRF1 acts against chilling stress in rice.